You can find the most updated list of publications here:
7.
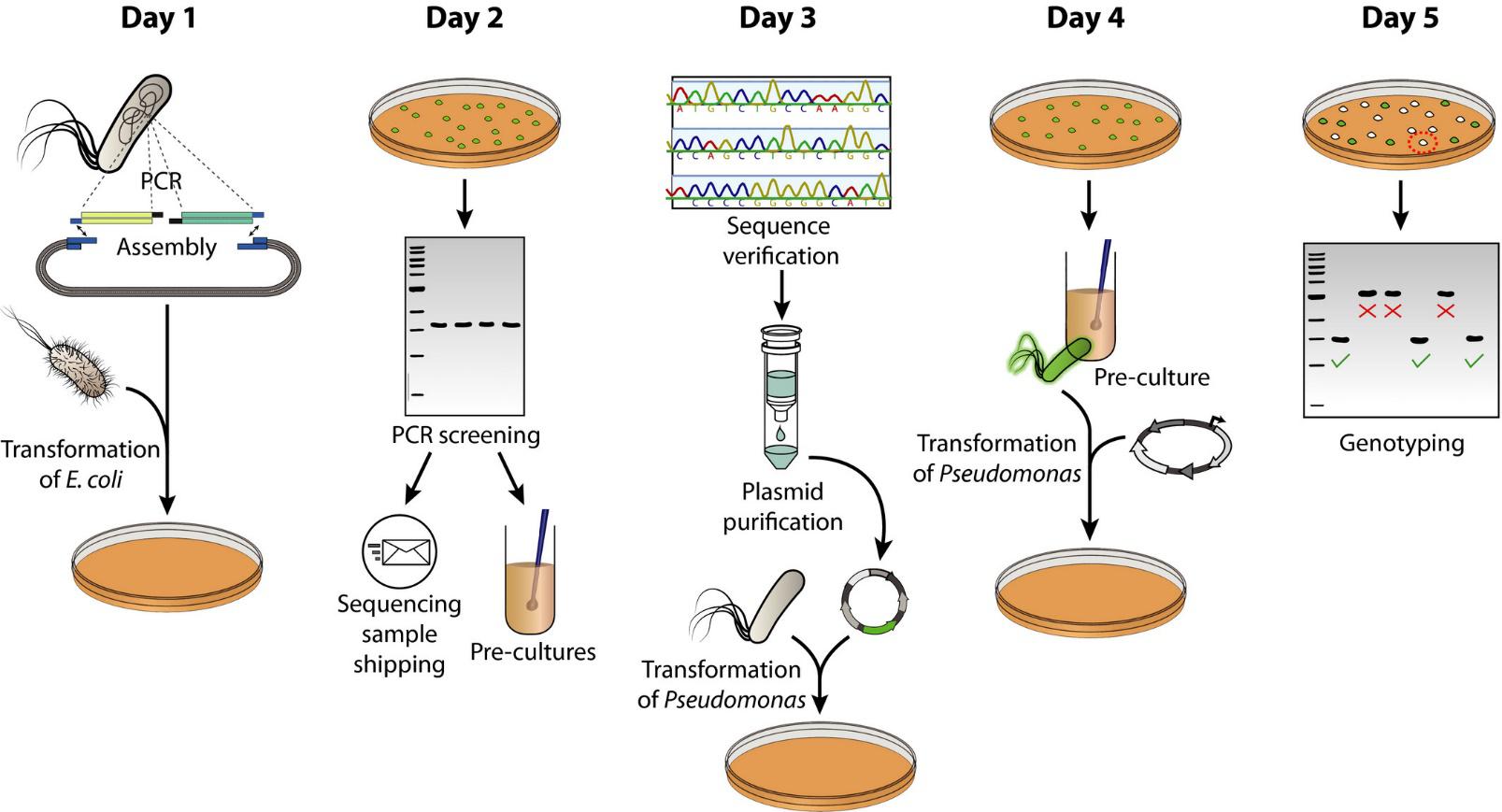
Wirth, N. T., Kozaeva, E., and Nikel, P. I. (2019) Accelerated genome engineering of Pseudomonas putida by I‐SceI―mediated recombination and CRISPR‐Cas9 counterselection. Microbial Biotechnology, 13: 233-249. DOI: 10.1111/1751-7915.13396
Speeding-up genetic manipulations in bacteria is a constant challenge in the pipeline of developing microbial cell factories. In this article, Wirth et al. shortened the time for deleting and integrating genomic DNA in P. putida to 3-5 days by improving screening methods and counterselection strategies.
Visit Publication6.
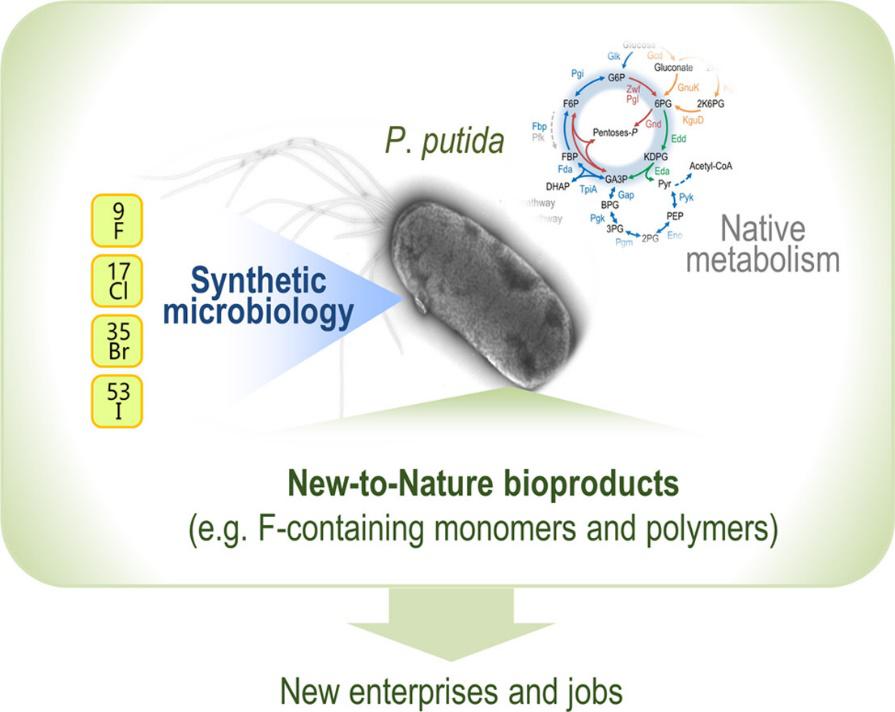
Martinelli, L., and Nikel, P. I. (2019) Breaking the state‐of‐the‐art in the chemical industry with new‐to‐Nature products via synthetic microbiology. Microbial Biotechnology, 12:187-190. DOI: 10.1111/1751-7915.13372
What if we could synthesize chemicals containing non-biological elements, such as halogens, using microbial cell factories? The EU Horizon2020—funded project SinFonia explores innovative possibilities to revolutionize the fluorine chemical industry using metabolic engineering and synthetic biology in P. putida.
Visit Publication5.
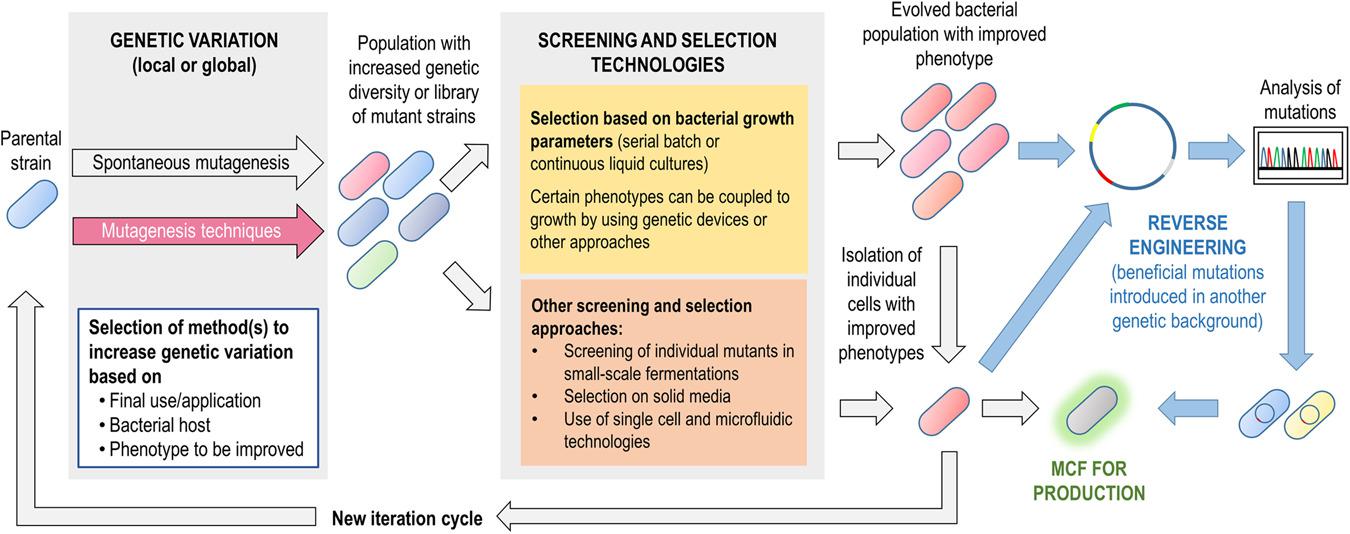
Férnandez-Cabezón, L., Cros, A., and Nikel, P. I. (2019) Evolutionary approaches for engineering industrially relevant phenotypes in bacterial cell factories. Biotechnology Journal, 14: e1800439. DOI: 10.1002/biot.201800439
Strategies involving bacterial evolution and genetic variability are of great importance in order to tackle the challenges in designing and constructing efficient cell factories. Different techniques, screening methods, and future trends are thoroughly covered in this review by Lorena, Antonin and Pablo.
Visit Publication4.
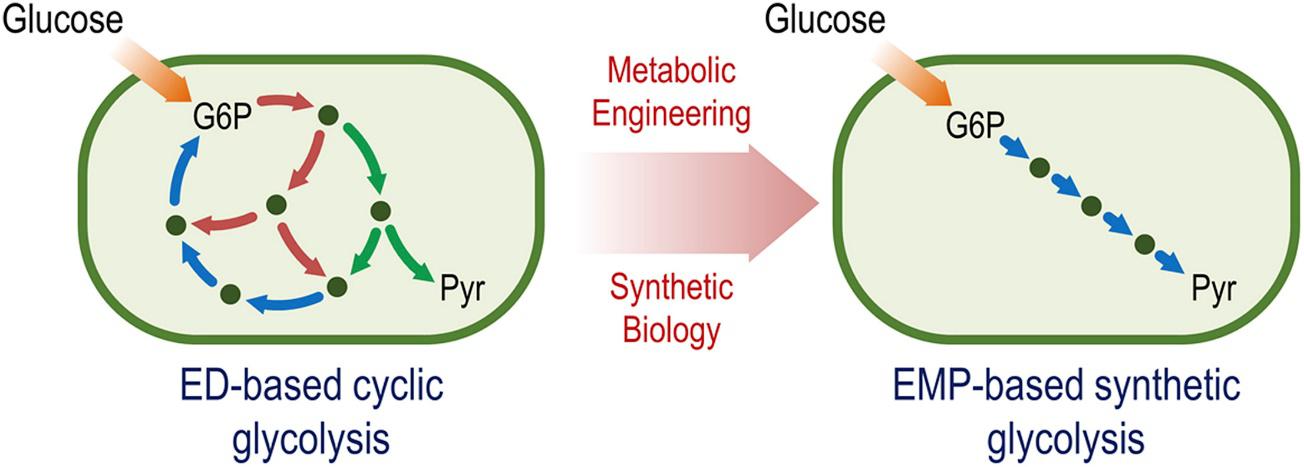
Sánchez-Pascuala, A., Férnandez-Cabezón, L., de Lorenzo, V., and Nikel, P. I. (2019) Functional implementation of a linear glycolysis for sugar catabolism in Pseudomonas putida. Metabolic Engineering, 54: 200-211. DOI: 10.1016/j.ymben.2019.04.005
Pseudomonas species run a cyclic, Entner-Doudoroff–based glycolysis, which allows them to adjust the availability of NAD(P)H depending on the external conditions. In this article, the native cyclic glycolysis of P. putida is rewired into a linear, Embden-Meyerhof-Parnas–based catabolism, which, upon further engineering of the host, led to improvements in carotenoid synthesis.
Visit Publication3.
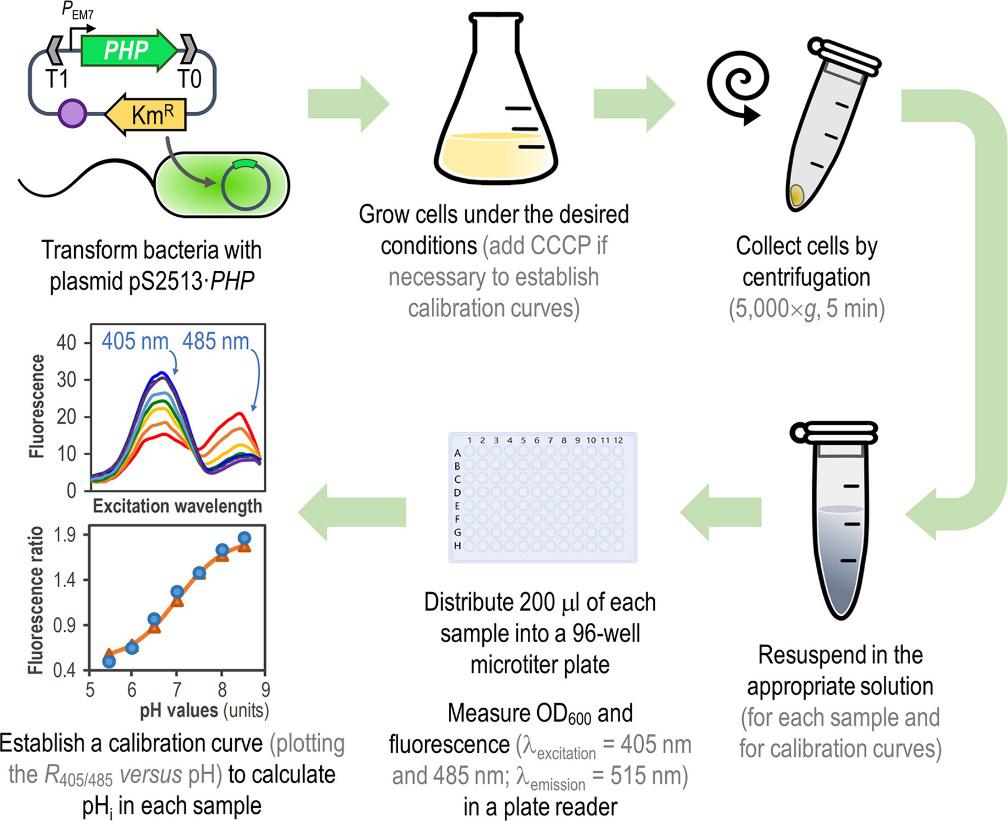
Arce-Rodríguez, A., Volke, D. C., Bense, S., Häussler, S., and Nikel, P. I. (2019) Non‐invasive, ratiometric determination of intracellular pH in Pseudomonas species using a novel genetically encoded indicator. Microbial Biotechnology, 12: 799-813. DOI: 10.1111/1751-7915.13439
In this research article, Arce-Rodríguez et al. developed and describe a fluorescence-based biosensor for assessing the intracellular pH of Gram-negative bacteria (with E. coli, P. putida and P. aeruginosa as examples), which helps to track changes in the pH in cells due to both internal and external changes.
Visit Publication2.
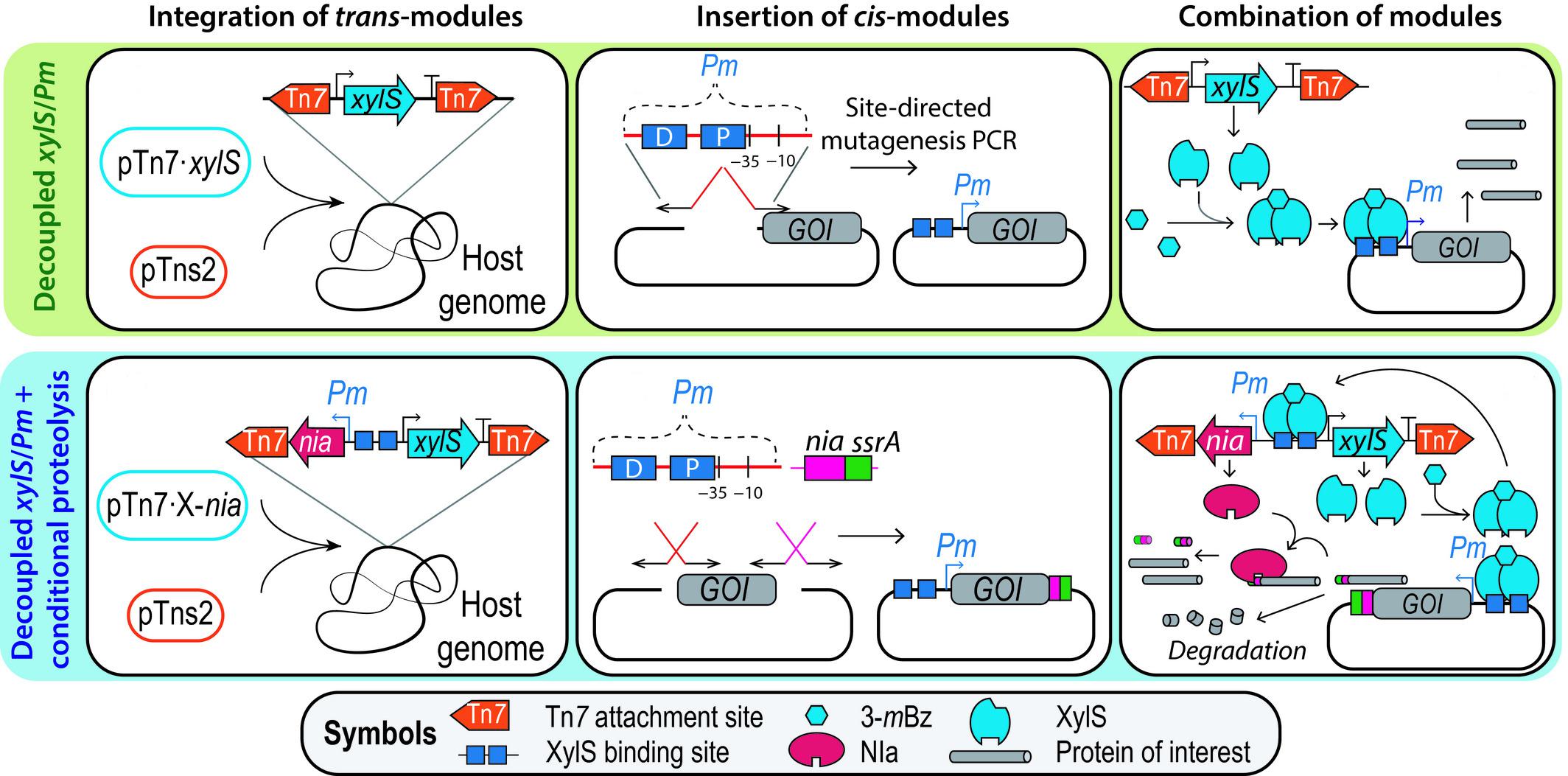
2. Volke, D. C., Turlin, J., Mol, V., and Nikel, P. I. (2019) Physical decoupling of XylS/Pm regulatory elements and conditional proteolysis enable precise control of gene expression in Pseudomonas putida. Microbial Biotechnology, 13: 222-232. DOI: 10.1111/1751-7915.13383
Optimizing expression systems is a key aspect in the design of cell factories. For this reason, Volke et al. improved the performance of the widely used XylS/Pm expression system in P. putida by integrating the transcriptional regulator XylS into the genome, and adding a proteolytic tag to the protein of interest to minimize the impact of leaky (basal) expression of the cognate gene.
Visit Publication1.
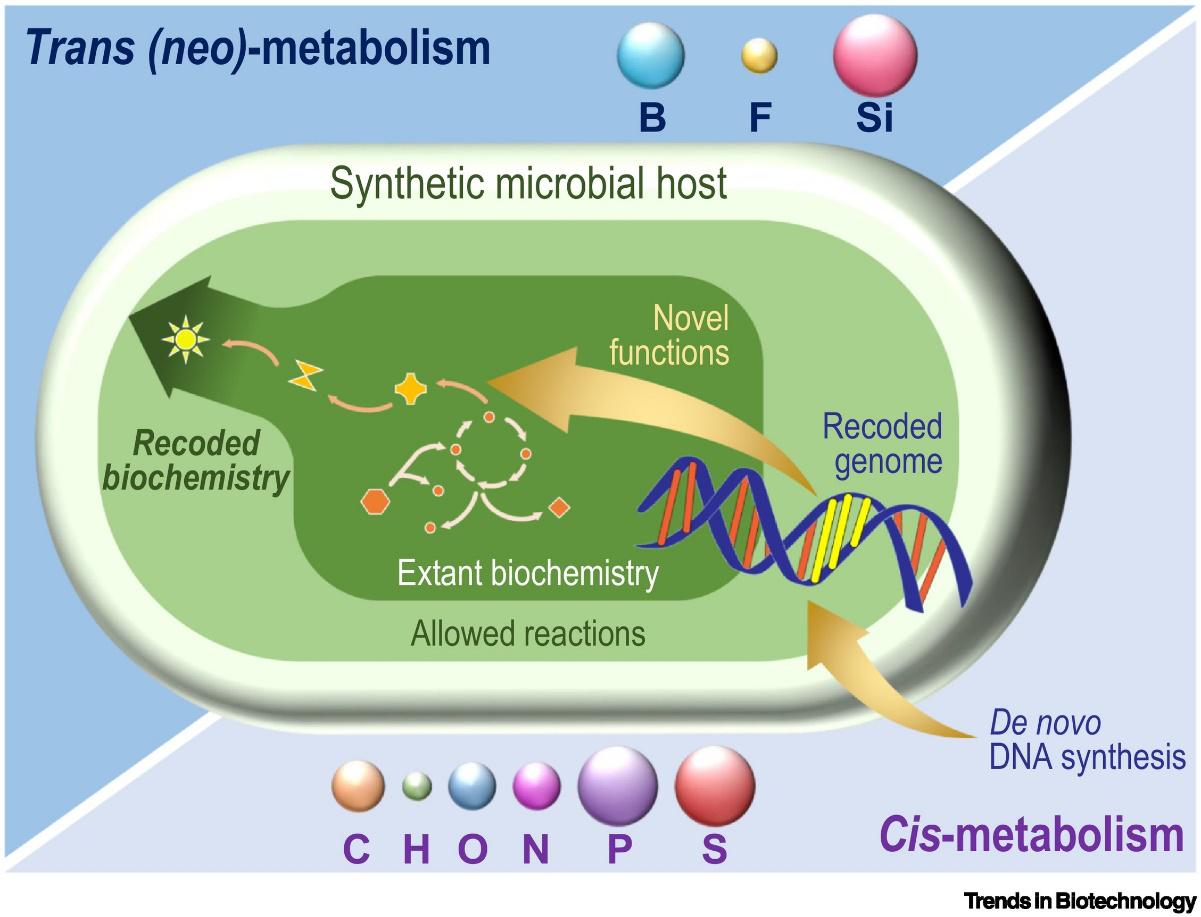
Nikel, P. I. (2019) Synthesis of recoded bacterial genomes toward bespoke biocatalysis. Trends in Biotechnology, 37: 1036-1038. DOI: 10.1016/j.tibtech.2019.07.001
In this spotlight article, Pablo assess the importance and future implications of constructing synthetic organisms after the ground-breaking work of Fredens et al. recoding the genome of Escherichia coli.
Visit Publication