You can find the most updated list of publications here:
10.
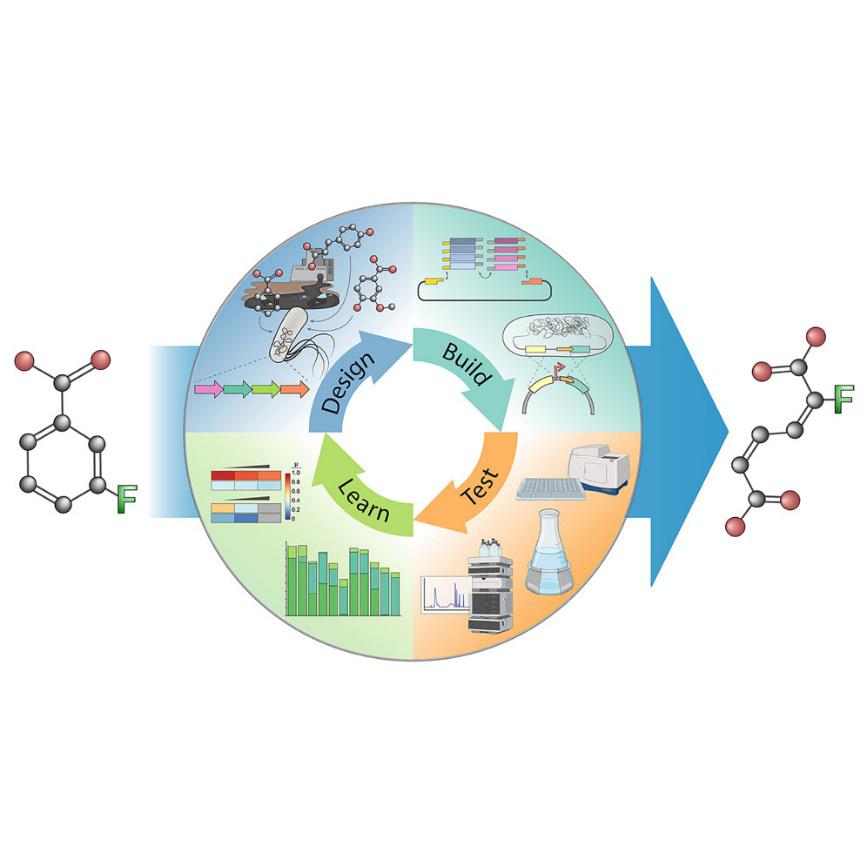
Wirth, N. T., and Nikel, P. I. (2021) Combinatorial pathway balancing provides biosynthetic access to 2-fluoro-cis,cis-muconate in engineered Pseudomonas putida. Chem Catalysis, https://doi.org/10.1016/j.checat.2021.09.002.
In this article, Nico Wirth and Pablo exploited the natural metabolism of P. putida to produce the new-to-industry halogenated molecule 2-fluoro-cis,cis-muconate (a chemical building block) from fluorinated benzoates. By gaining a deep understanding of the transcriptional and metabolic imbalances within the conversion pathway, they managed to reach the theoretical yield of the product of interest!
Visit Publication9.
Raman, K., Sinha, H., Vickers, C. E., and Nikel, P. I. (2021) Synthetic Biology beyond borders. Microbial Biotechnology, DOI: 10.1111/1751-7915.13966
This is an editorial for the special issue of the same name in Microbial Biotechnology, where the advances in engineering biology as well as the current challenges of Synthetic Biology for solving global problems are discussed.
Visit Publication8.
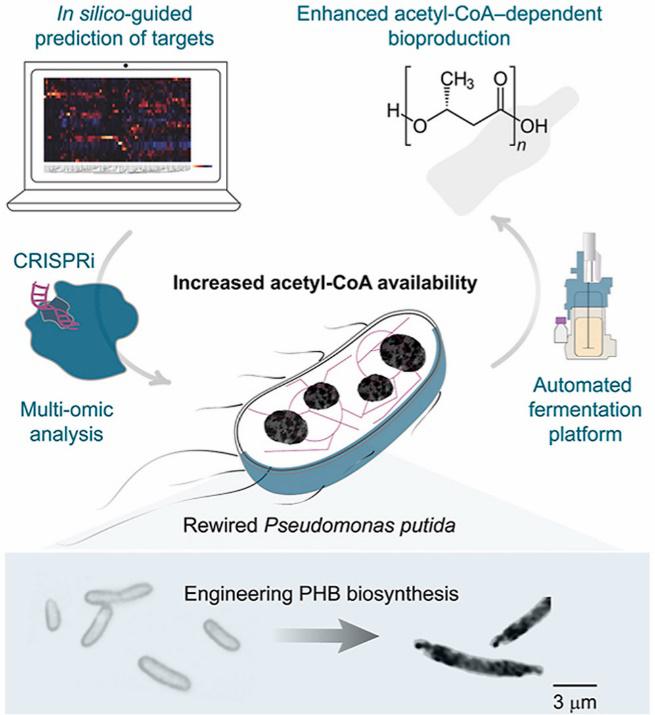
Kozaeva, E., Volkova, S., Matos, M. R. A., Mezzina, M. P., Wulff, T., Volke, D. C., Nielsen, L. K., and Nikel, P. I. (2021) Model-guided dynamic control of essential metabolic nodes boosts acetyl-coenzyme A–dependent bioproduction in rewired Pseudomonas putida. Metabolic Engineering, https://doi.org/10.1016/j.ymben.2021.07.014
A combination of in silico predictions, and pathway and cell morphology engineering through the implementation of a CRISPR toolset was used to increase the availability of acetyl-coenzyme A in P. putida. Acetyl-CoA is a key building block for multiple bioproduction pathways and, as an example, the production of biopolymers (polyhydroxyalkanoates) was demonstrated in this research article by Ekaterina.
Visit Publication7.
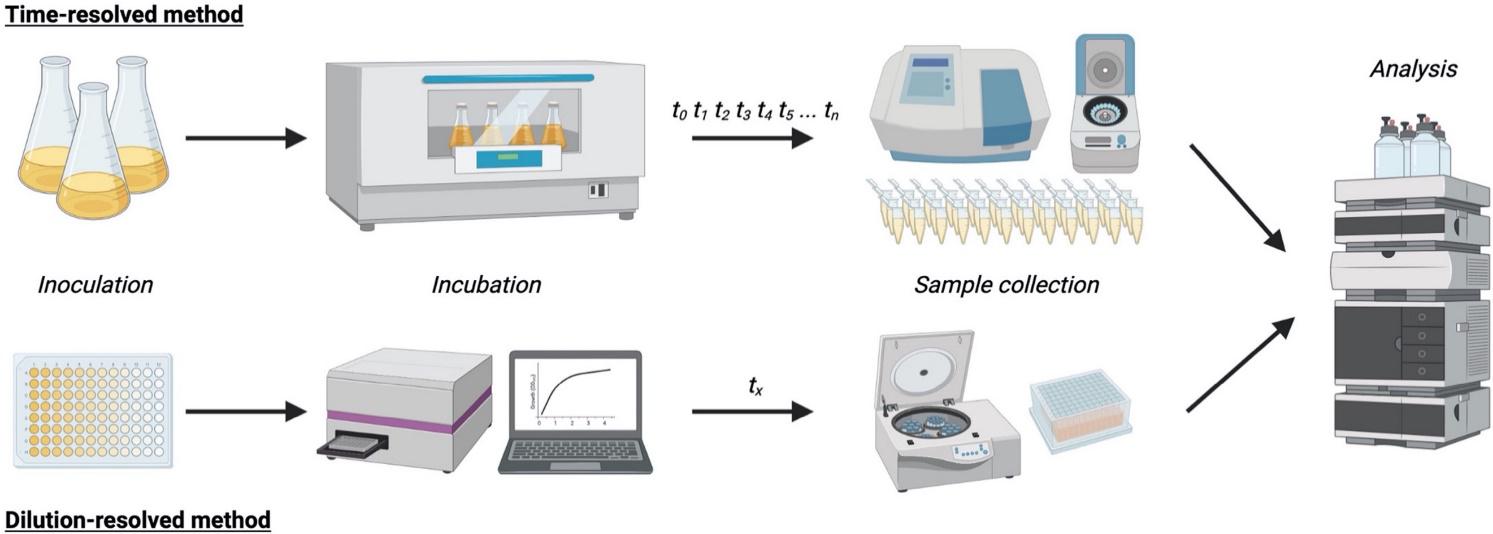
Pedersen, B. H., Gurdo, N., Johansen, H. K., Molin, S., Nikel, P. I., and La Rosa, R. (2021) High-throughput dilution-based growth method enables time-resolved exo-metabolomics of Pseudomonas putida and Pseudomonas aeruginosa. Microbial Biotechnology, DOI: https://doi.org/10.1111/1751-7915.13905
In this paper, part of a collaboration with Prof. S. Molin’s laboratory, Nico Gurdo developed an innovative high-throughput method for metabolomic footprint of P. putida and P. aeruginosa strains using dilution-resolved cultivation in 96-deep well plates.
Visit Publication6.
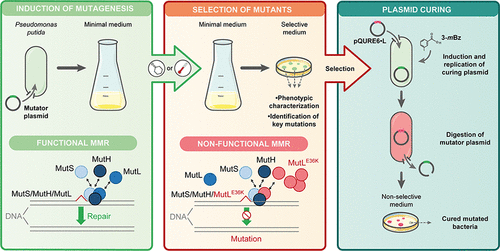
Fernández-Cabezón, L., Cros, A., and Nikel, P. I. (2021) Spatiotemporal manipulation of the mismatch repair system of Pseudomonas putida accelerates phenotype emergence. ACS Synthetic Biology, DOI: https://doi.org/10.1021/acssynbio.1c00031
The evolution of different phenotypes, such as antibiotic resistances and synthetic auxotrophies, was improved in this research programme led by Lorena and Antonin. Using a conditional induction system to interfere with the mismatch repair system in P. putida, they managed to create strains with potential for fast Adaptive Laboratory Evolution (ALE).
Visit Publication5.
Nikel, P. I., and Mattanovich, D. (2021) Microbial cell factories: a biotechnology journey across species. Essays in Biochemistry, DOI: https://doi.org/10.1042/EBC20210037
In this editorial of Essays in Biochemistry, the authors present a curated collection of reviews of different traditional microbial cell factories, as well as promising new chassis organisms.
Visit Publication4.
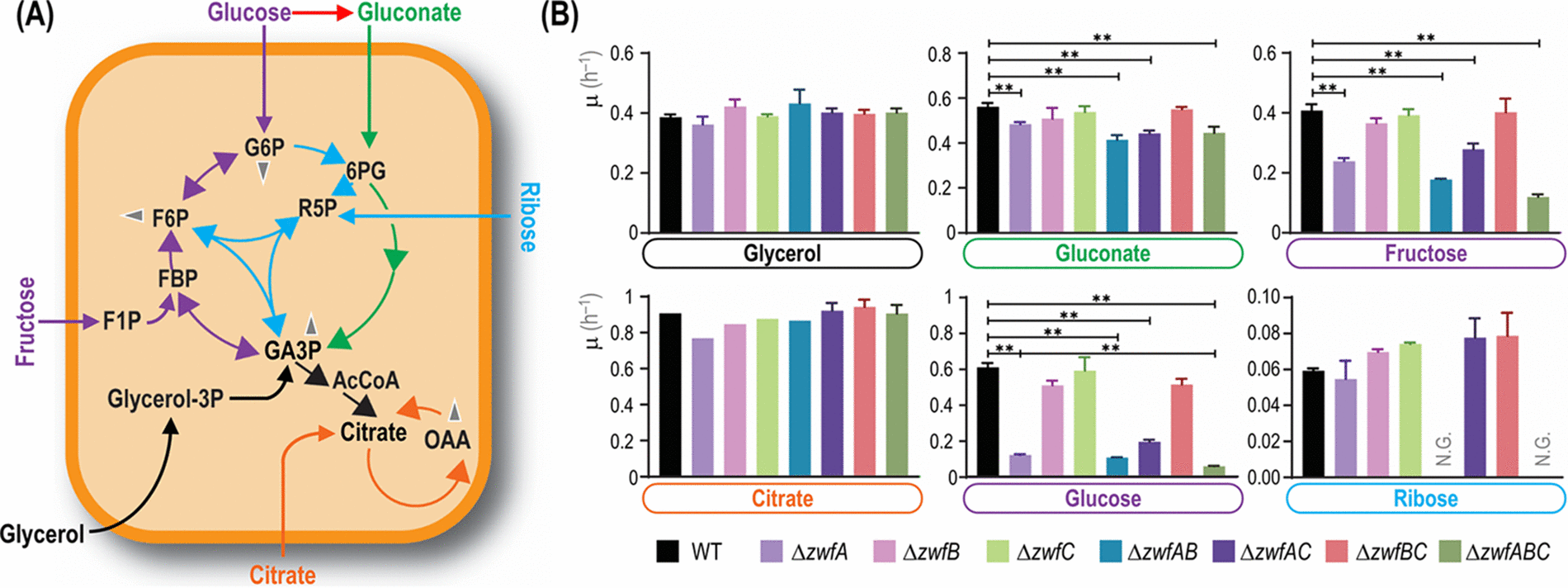
Volke, D. C., Olavarría, K., and Nikel, P. I. (2021) Cofactor specificity of glucose-6-phosphate dehydrogenase isozymes in Pseudomonas putida reveals a general principle underlying glycolytic strategies in bacteria. mSystems, DOI: https://doi.org/10.1128/mSystems.00014-21
In this study led by Daniel, the evolutionarily-defined role of glucose-6-phosphate dehydrogenase isozymes was exposed by assessing the enzymes’ specificities towards NADP+ and NAD+ and their impact on the global redox balance of P. putida.
Visit Publication3.
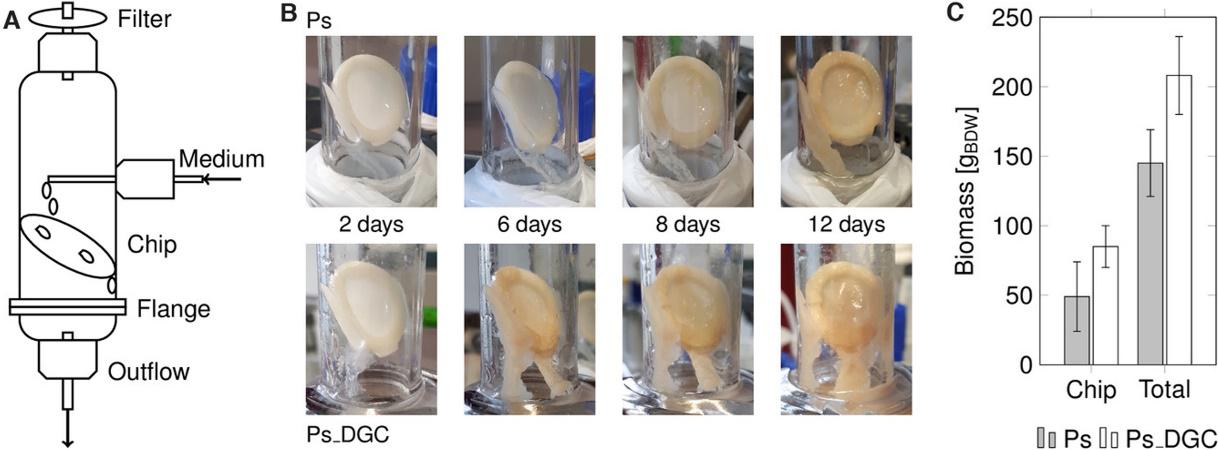
Heuschkel, I., Hanisch, S., Volke, D. C., Löfgren, E., Hoschel, A., Nikel, P. I., Karande, R., and Bühler, K. (2021) Pseudomonas taiwanensis biofilms for continuous conversion of cyclohexanone in drip flow and rotating bed reactors. Engineering in Life Sciences, DOI: doi.org/10.1002/elsc.202000072
Different types of bioreactors for biofilm formation were tested for engineered P. taiwanensis. The strains used in this article, which efficiently convert cyclohexanone to ε-caprolactone, were also re-wired to generate thick synthetic biofilms, improving the overall conversion process.
Visit Publication2.
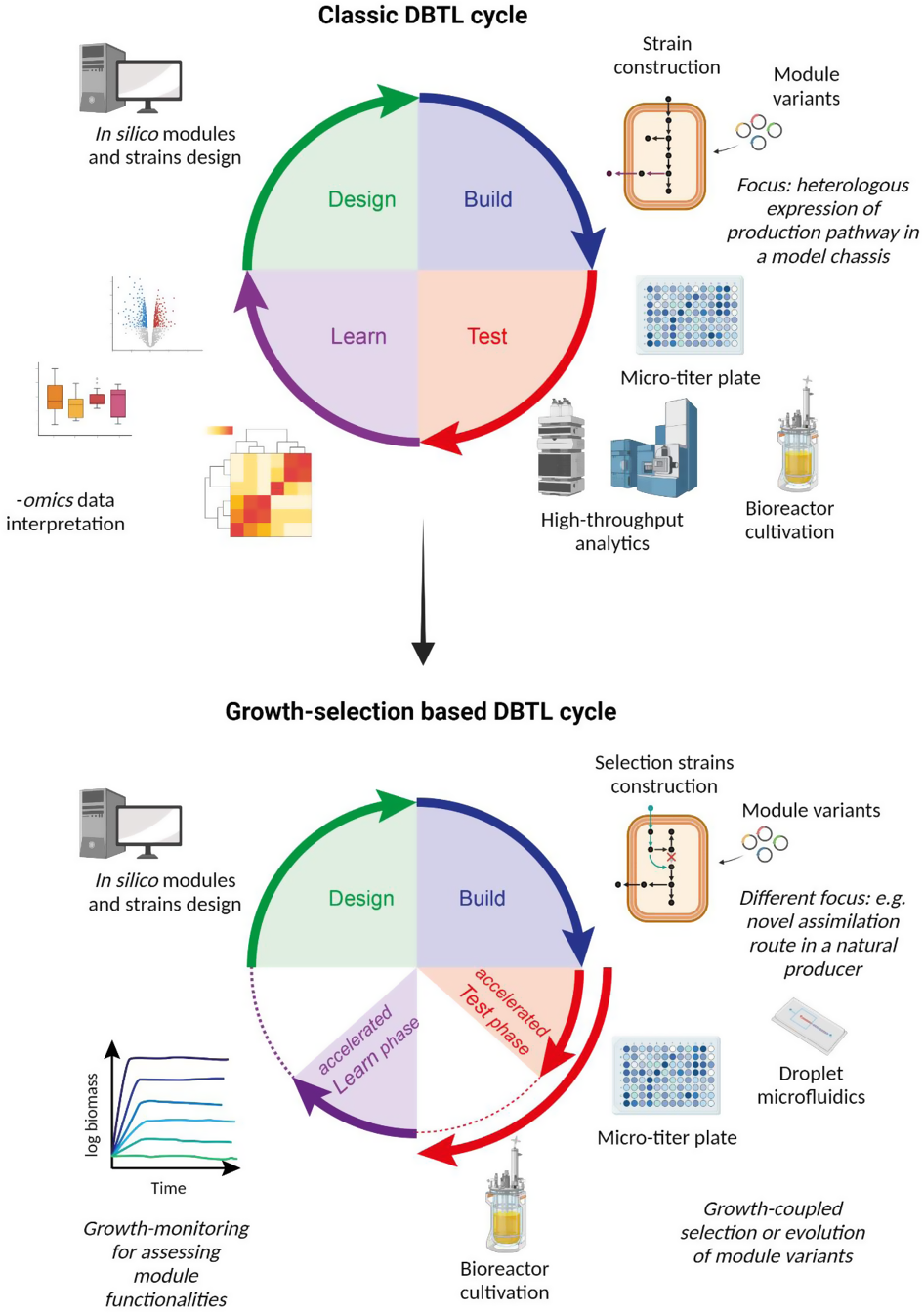
Orsi, E., Claassens, N. J., Nikel, P. I., and Lindner, S. (2021) Growth-coupled selection of synthetic modules to accelerate cell factory development. Nature Communications, doi.org/10.1038/s41467-021-25665-6
In this opinion article, the authors discuss the potential of using strategies to improve enzyme or pathway activities by forcing bacteria to grow using such (synthetic) activities. Therefore, under pressure conditions, an increase in the growth would lead to an increase in the production of target compound(s)!
Visit Publication1.
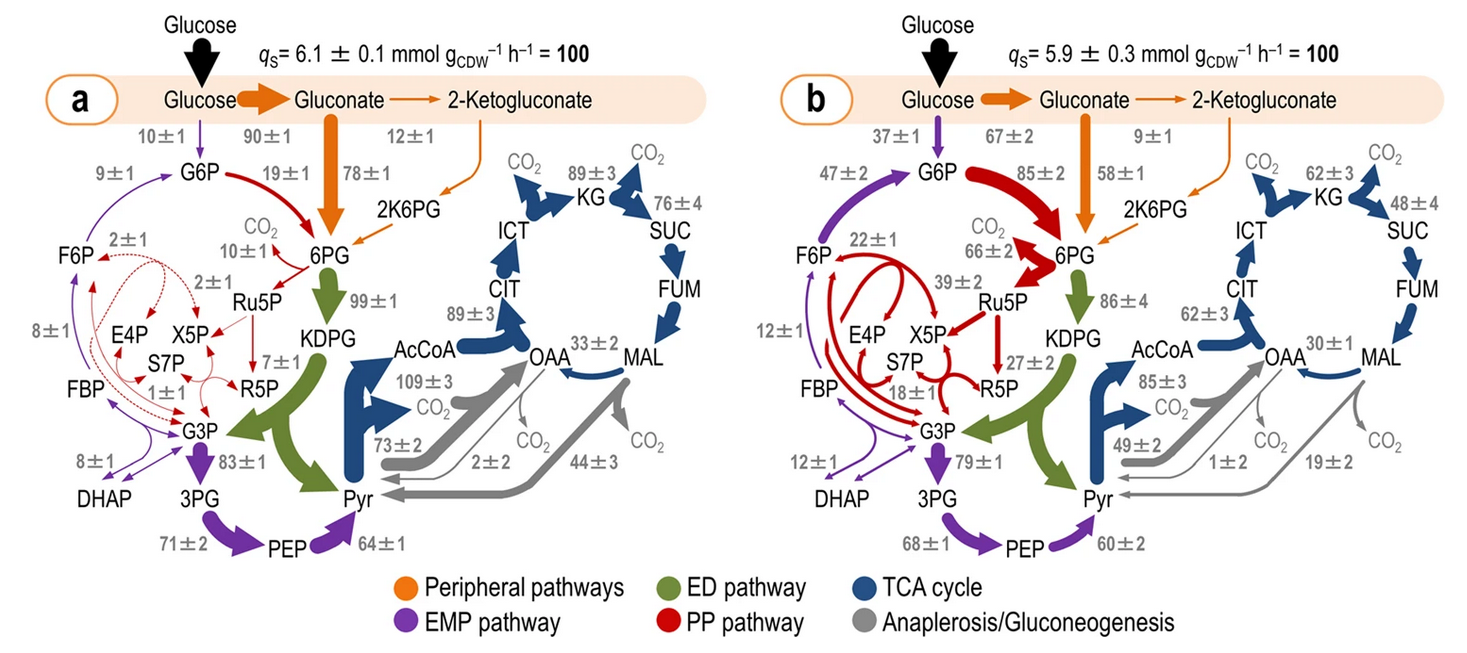
Nikel, P. I., Fuhrer, T., Chavarría, M., Sánchez-Pascuala, A., Sauer, U., and de Lorenzo, V. (2021) Reconfiguration of metabolic fluxes in Pseudomonas putida as a response to sub-lethal oxidative stress. The ISME Journal, DOI: https://doi.org/10.1038/s41396-020-00884-9
This article describes how the carbon fluxes in central metabolism undergo a deep reorganization when the cells are exposed to oxidative insults—similar to the conditions that P. putida encounters in its typical niches. This reconfiguration results in a significant increase in the fluxes that generate NADPH, the metabolic currency key to counteract reactive oxidative species.
Visit Publication